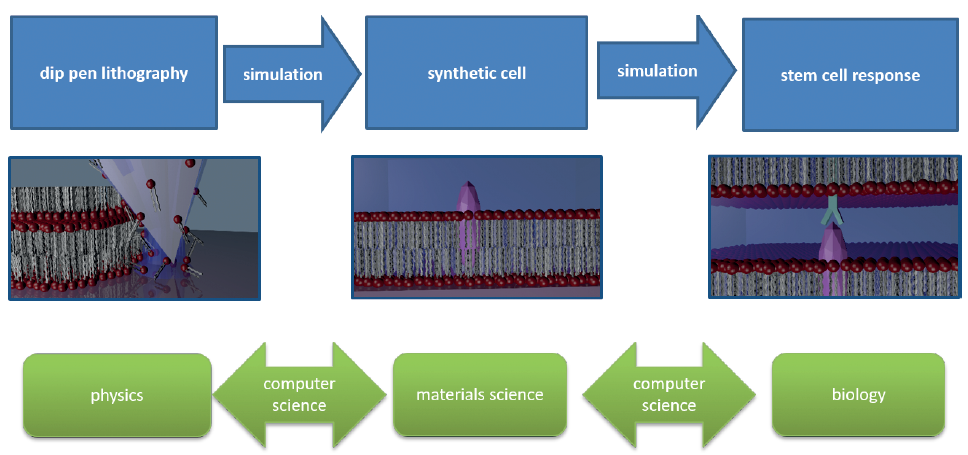
Bringing together simulation and experiment in lithography with lipids
Last year, we started our endeavor for a better understanding on the pro cesses in writing phospholipid struc tures and how the resulting synthetic biomimetic membranes interact with and instruct cells. The YIN Grant en abled us to do piloting studies focus ing on the writing process via dip-pen nanolithography with phospholipids (L-DPN) – the first step in modeling the full chain of processes from writing to cell-surface interaction.
The generation of bioactive materials and interfaces usually involves highly interdisci plinary work. Here we engage in (i) physics for creating lipid membranes by dip-pen nanolithography (DPN) with phospholipids, (ii) material science for reconfiguring these membranes in liquid and incorporating sig nal protein structures, and (iii) biology to study the interaction of cells (on molecular level) with these structures.
For each step, there are still significant knowl edge gaps concerning the exact mechanisms and processes which are not readily accessible with experiments. Here, computer simulations can help as “computational microscope” to shed light on the blind spots. Atomistic simulations allow to observe the underlying microscopic mechanisms, which, in turn, make it possible to formulate the design strategies for tailored and printed substrates.
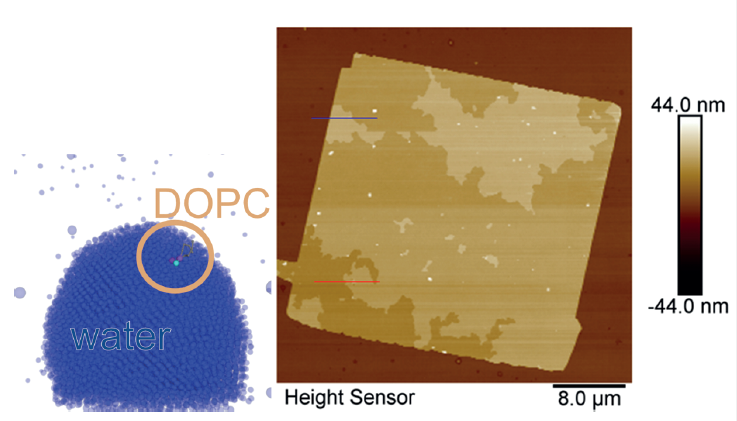
We focused on writing complex molecules like the phospholipids via DPN: For the first time, we used a simulation including tip, meniscus and phospholipid molecules to catch a microscopic view on the membrane writing process. With the focus on atomic scale process-structure relation, coarse grained molecular dynamics visualized the migration of the lipids towards the substrate in a simplified DPN environment. First, the water-lipid interaction was simulated on a drop let wetted substrate, in order to separate the col lective bilayer membrane formation from the lipid diffusion on water.
With the initial results from the grant period, we established simulation tools that can give first correlations between simulation and experiment.